Superfund Research Program
Leveraging Machine Learning to Predict Toxicity
View Research Brief as PDF(428KB)
Release Date: 03/02/2022
![]() subscribe/listen via iTunes, download(6.6MB), Transcript(88KB)
subscribe/listen via iTunes, download(6.6MB), Transcript(88KB)
NIEHS Superfund Research Program (SRP) grantees developed a new computational approach to predict how hazardous substances may affect health based on key changes in cells. Led by April Z. Gu, Ph.D., of the Northeastern University Puerto Rico Testsite for Exploring Contamination Threats (PROTECT) SRP Center, researchers used machine learning and advanced algorithms to link biological changes from high throughput cell studies with health outcomes observed in animal studies.
The team also included former PROTECT trainee Sheikh Mokhlesur Rahman, Ph.D., and investigators David Kaeli, Ph.D., and Akram Alshawabkeh, Ph.D., from Northeastern University SRP Center.
Identifying the Most Relevant Biomarkers
By identifying and prioritizing a few key biomarkers, the team aimed to reduce complexity in traditional toxicity screening methods, which are costly and time-consuming. Biomarkers are molecules - such as proteins, genes, or hormones - that indicate biological changes in cells and are used to measure the presence or progress of disease.
Researchers conducted two case studies to test whether their identified biomarkers could predict health outcomes. The first focused on biomarkers of cancer in rodents, and the second looked at genetic toxicity in bacteria. In both case studies, the most widely used biomarkers by government agencies, such as the National Toxicology Program (NTP), were tested to evaluate the ability of chemicals to cause genetic changes that may lead to cancer.
For each case study, they used data on six concentrations of 13 chemicals reported to cause cancer and a control group of 7 chemicals that do not cause cancer. A computational approach was used to evaluate a library of 38 proteins involved in DNA damage and repair activities. Changes to these proteins can lead to severe DNA damage and to mutations in genes that increase the likelihood of tumor formation and diseases, such as cancer.
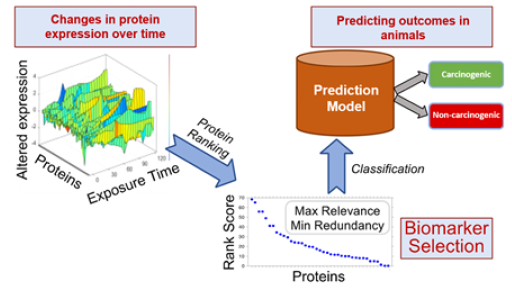
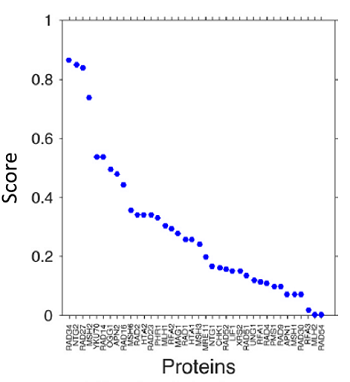
Using an algorithm called maximum relevance minimum redundancy, they selected which biomarkers were more closely associated with carcinogenic chemicals, such as formaldehyde and benzo[a]pyrene. Then, they narrowed down the list by identifying which biomarkers were redundant, associated with the same chemicals and biological mechanisms. Removing redundant biomarkers allowed the researchers to decrease the size and complexity of their database.
This approach identified five out of the 38 proteins, NTG2, RAD34, RAD27, MSH2, and YKU70, as the most relevant biomarkers in rodents, and APN2, RFA2, NTG2, RAD2, and MSH6 as the best predictive markers for genetic toxicology studies using bacteria.
Evaluating Performance
To assess the performance of these biomarkers to predict carcinogenic and genetic toxicity they applied a classification algorithm, called support vector machine to the case studies. Compared to other classification algorithms, this approach can reliably classify chemicals while avoiding overfitting and reducing susceptibility to noisy or meaningless data. Overfitting is an error that occurs when the machine learning model is too closely related to the dataset, and therefore loses its applicability to any other dataset.
Using only the top five biomarkers, the team reported 76% accuracy in classifying and predicting chemicals that cause cancer in rodents and 70% accuracy in predicting genetic toxicity in bacteria. When all 38 proteins were included in the prediction model, the accuracy was increased to 83% for the rodent case study and 78% for bacteria.
According to the authors, although the full library of biomarkers may yield slightly better prediction scores, the top-ranked five biomarkers can achieve relatively high prediction accuracy while reducing the cost, time, and complexity of screening chemicals for further study.
Their findings have important implications for achieving the goals of the Tox21 initiative, a federal collaboration that is developing alternative, non-animal methods to quickly and efficiently test thousands of chemicals for potential health effects.
For More Information Contact:
April Z Gu
Cornell University
263 Hollister Hall
Ithaca, New York 14850
Phone: 607-255-8778
Email: aprilgu@cornell.edu
To learn more about this research, please refer to the following sources:
- Rahman SM, Lan J, Kaeli D, Dy J, Alshawabkeh AN, Gu AZ. 2022. Machine learning-based biomarkers identification from toxicogenomics - bridging to regulatory relevant phenotypic endpoints. J Hazard Mater 423:doi:10.1016/j.jhazmat.2021.127141 PMID:34560480
To receive monthly mailings of the Research Briefs, send your email address to srpinfo@niehs.nih.gov.


