Superfund Research Program
Microbial Biogeochemical Cycling of Arsenic and of Chromium Coupled to the Biodegradation of Aromatic Contaminant Compounds
Project Leader: Lily Y. Young (Rutgers University)
Grant Number: P42ES010344
Funding Period: 2000 - 2006
Project-Specific Links
- Project Summary
Final Progress Reports
Year: 2005
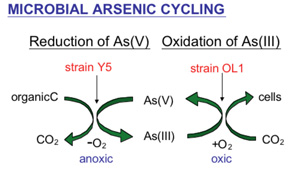
Naturally occurring arsenic is found predominantly as arsenate [As(V)] or arsenite [As(III)], and can readily be oxidized or reduced by microorganisms. Given the health risks associated with arsenic in groundwater and the interest in arsenic-active microorganisms, Dr. Young and her research team hypothesized that environmental microorganisms could mediate a redox cycling of arsenic that is linked to their metabolism. This hypothesis was tested using an As(V) respiring reducer (strain Y5) and an aerobic chemoautotrophic As(III) oxidizer (strain OL1) both isolated from a Superfund site, Onondaga Lake, in Syracuse, NY. Strains were grown separately and together in sealed serum bottles, and the oxic/anoxic condition was the only parameter changed. Initially, under anoxic conditions when both isolates were grown together, 2mM As(V) was stoichiometrically reduced to As(III) within 14 days. Following complete reduction, sterile ambient air was added and within 24 hours As(III) was completely oxidized to As(V). The anoxic-oxic cycle was repeated, and sterile controls showed no abiotic transformation within the 28-day incubation period. These results demonstrate that microorganisms can cycle arsenic in response to dynamic environmental conditions and thereby affecting the speciation, and hence mobility and toxicity of arsenic in the environment.
Autotrophic microorganisms have been isolated that are able to derive energy from the oxidation of arsenite [As(III)] to arsenate [As(V)] under aerobic conditions. Based on chemical energetics, microbial oxidation of As(III) can occur in the absence of oxygen, and may be relevant in some environments. Enrichment cultures were established from an arsenic contaminated industrial soil amended with As(III) as the electron donor, inorganic C as the carbon source and nitrate as the electron acceptor. In the active enrichment cultures, oxidation of As(III) was stoichiometrically coupled to the reduction of NO3-. Two autotrophic As(III) oxidizing strains were isolated that completely oxidized 5mM As(III) within 7 days under denitrifying conditions. Based on 16S rRNA gene sequencing results, strain DAO1 was 99% related to Azoarcus and strain DAO10 was most closely related to a Sinorhizobium. The nitrous oxide reductase (nosZ) and the RuBisCO Type II (cbbM) genes were successfully amplified from both isolates underscoring their ability to denitrify and fix CO2 while coupled to As(III) oxidation. Although limited work has been done to examine the diversity of anaerobic autotrophic oxidizers of As(III), this process may be an important component in the biological cycling of arsenic within the environment.
Table 2. Nitrate utlization is stoichiometric to arsenic oxidation in the original culture and in each of the 2 pure cultures under denitrifying conditions. The amount of nitrate used closely matches that which is predicted. The only added carbon source was carbonate/bicarbonate. Stoichiometry was based on the following:
0.5 H3AsO30 + 0.2NO3- --> 0.5HAsO42- + 0.1N2 + 0.8H+ + 0.1H2O
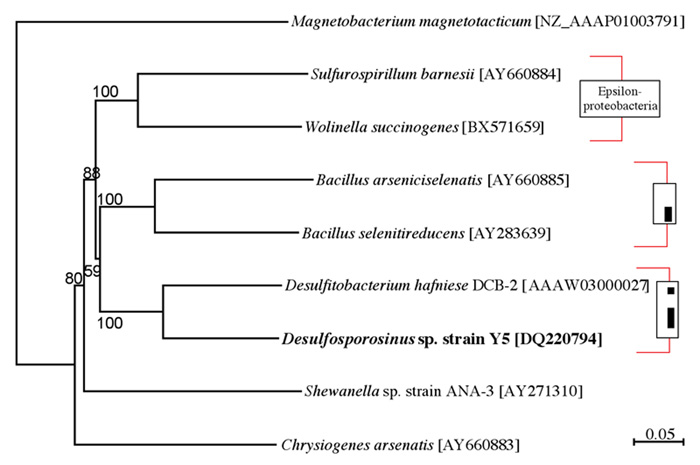
Desulfosporosinus sp. strain Y5 is a spore-forming bacterium capable of dissimilatory arsenate reduction coupled to the oxidation of aromatic compounds. In arsenate respiration, the arsenate respiratory reductase (ARR) catalyzes the reduction of arsenate to arsenite. The project investigator's objective is to characterize the arrA gene, encoding the ARR, for Desulfosporosinus sp. strain Y5. Oligonucleotide primers were designed based on the few arrA gene sequences available at the time, and validated against positive and negative controls. The resulting arrA-amplicon of ~2.0 kb was cloned and sequenced. The arrA from Desulfosporosinus sp. Y5 is closely related to Desulfitobacterium hafniense (similarity of 77% and 81% at the nucleotide and amino acid levels, respectively). Phylogenetic topology based on the arrA gene was partially congruent with that of 16S rRNA-based analysis. This arrA sequence will support the development of specific tracking probes for Desulfosporosinus sp. Y5 and the molecular characterization and monitoring of dissimilatory arsenate reducing bacteria.
Fig. 2. Phylogenetic relationship of arsenate-respiring bacteria based on arrA gene sequences. The tree was reconstructed using neighbor-joining method with bootstrap of 1000 repetitions on ~2.0 kb fragments. The percentage values for bootstrap is indicated to be found in their habitats.
NOTE:
Superfund site used in study include Onondaga Lake, Syracuse, NY.