Superfund Research Program
Improved Sequencing Method Leads to Advancements in Toxicology Research
View Research Brief as PDF(657KB)
Release Date: 12/02/2020
![]() subscribe/listen via iTunes, download(6.6MB), Transcript(87KB)
subscribe/listen via iTunes, download(6.6MB), Transcript(87KB)
NIEHS-funded Superfund Research Program (SRP) scientists are employing a new RNA sequencing method to assess mechanisms of toxicity on a finer and more accessible scale. Researchers in SRP grantee Tim Zacharewski’s Lab at the Michigan State University (MSU) SRP Center conducted the study.
Using single-nuclei RNA sequencing (snSeq), lead researcher Rance Nault and the team were able to identify distinct hepatic cell types, cell population shifts and relative abundances, and discriminated cell-specific pathways disrupted by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD).
TCDD is an environmental contaminant produced from waste incineration, metal production, and fossil-fuel and wood combustion. TCDD is a potential contributing factor to complex metabolic diseases such as obesity, type II diabetes, and nonalcoholic fatty liver disease.
With the conventional method, bulk RNA sequencing (RNA-seq), researchers would grind up whole pieces of tissue and measure the average levels of RNA for each gene in the sample. This method can mask the diversity of cell types in the tissue and misses how each type of cell responds to chemicals or drugs. The snSeq method used in this study offers more information at a higher resolution about how TCDD affects each individual cell type.
Finer Findings
Using frozen mouse liver samples treated with TCDD, the team compared snSeq data to historical RNA-seq datasets. The comparison showed that the snSeq technology can reproduce previous findings, in addition to providing cell-specific information. The researchers captured major shifts in cell populations from these gene expression data and produced cluster graphs to visualize the cell-specific expression.
Single-cell RNA sequencing (scSeq) is the most common sequencing approach and can only be performed on freshly collected tissues. In typical toxicology study designs multiple doses of chemicals, drugs, or supplements are used to establish their safety. The number of samples, coupled with the severe effects observed at higher doses, presents a significant hurdle in the use of freshly isolated single cells.
The MSU researchers were able to adapt a snSeq protocol for frozen cancer tissue biopsy samples from another study to complete their analysis. The team showed for the first time that by using nuclei isolated from frozen livers, it is possible to identify known responses caused by TCDD exposure, as well as gain novel insight into how specific cell populations respond.
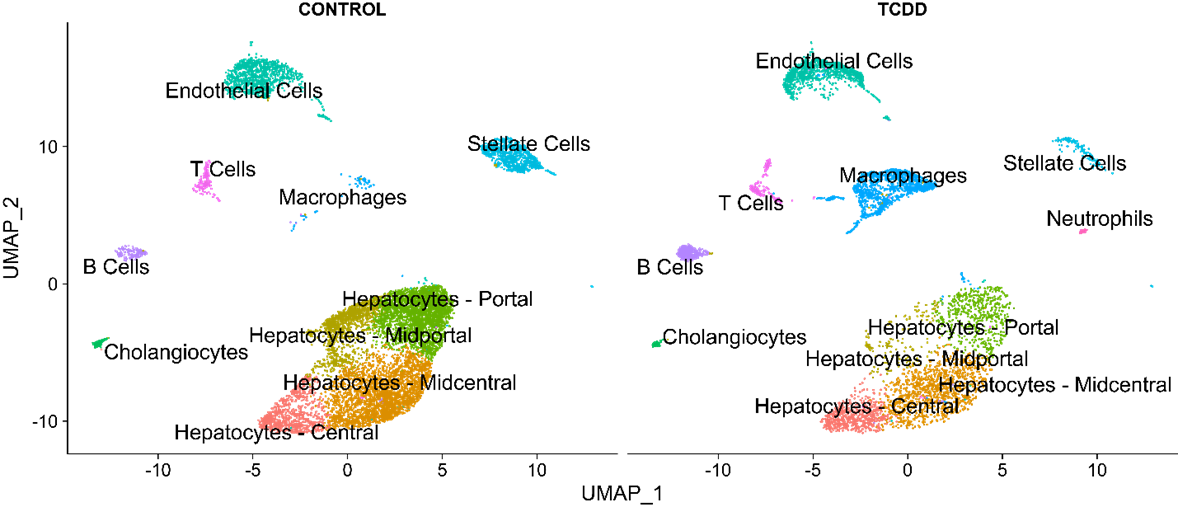
UMAP visualization shows clustering of individual nuclei based on the similarity in the expression of thousands of individual genes, represented as individual points, allowed the research team to characterize the diversity of distinct liver cell types. Application of snSeq identified all major known liver cell types and revealed shifts in their relative abundance which can be visualized by changes in the density between control (left) and TCDD (right) treatment groups.
Additionally, because the snSeq method can analyze gene expression at unprecedented resolution, the researchers were able to identify and characterize rare cell types. This discovery further clarifies the significance of tissue spatial organization. This finding is important as it has been established that drugs and toxicants elicit spatial (zonal) toxicities. Acetaminophen, for example, primarily affects the centrilobular region of the liver due to higher expression of certain enzymes.
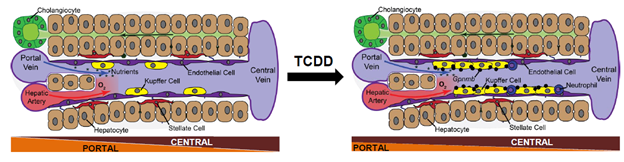
This illustration shows how snSeq technology was used to characterize the action of TCDD at the spatial, cellular, and molecular level in livers of mice exposed to TCDD. TCDD, acting as a portal toxicant, caused the infiltration and expansion of non-alcoholic steatohepatitis-associated macrophages.
Research Implications
Demonstration of the feasibility and value of a single-nuclei RNA sequencing approach was a critical step for applying it to more complex study designs. This improved mechanistic insight has wide-reaching implications in the field of toxicology and beyond. This method can be applied to other parts of the human system with different contaminants.
This work expands use of snSeq in toxicology. Research investigating environmental contaminants, drugs, and dietary supplements can use this methodology. Better targeted therapies and risk assessment, heightened knowledge of how to prevent toxicity and disease, and improved ability to set substance thresholds and limits are all potential outcomes.
Future efforts by Nault and the team of MSU SRP Center researchers will explore the use of this technology to characterize the cell-specific sensitivity upon exposure to liver toxicants in order to better understand the development and progression of toxicant-associated nonalcoholic fatty liver disease.
For More Information Contact:
Timothy R Zacharewski
Michigan State University
Department of Biochemistry and Molecular Biology
1129 Farm Lane, Room 241
East Lansing, Michigan 48824-1319
Phone: 517-355-1607
Email: tzachare@msu.edu
To learn more about this research, please refer to the following sources:
- Nault R, Fader KA, Zacharewski TR. 2021. Single-nuclei RNA sequencing assessment of the hepatic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Cell Mol Gastroenterol Hepatol 11:147-159. doi:10.1016/j.jcmgh.2020.07.012 PMID:32791302 PMCID:PMC7674514
To receive monthly mailings of the Research Briefs, send your email address to srpinfo@niehs.nih.gov.


