Superfund Research Program
First-of-its-Kind Arsenic Meta-Analysis Paves the Way for Future Data Integration
View Research Brief as PDF(426KB)
Release Date: 09/01/2021
![]() subscribe/listen via iTunes, download(7.5MB), Transcript(88KB)
subscribe/listen via iTunes, download(7.5MB), Transcript(88KB)
Researchers from NIEHS Superfund Research Program (SRP) centers at the University of California (UC), Berkeley and Columbia University used advanced analysis techniques to combine data from populations in Chile and Bangladesh. The purpose was to detect common DNA methylation (DNAm) signatures associated with arsenic exposure.
Arsenic is known to cause cancer in humans and is associated with a range of health problems including diabetes and cardiovascular disease. Epigenetic changes, like DNAm, can alter gene expression without directly altering DNA sequences. These changes might serve as biomarkers of exposure to chemicals like arsenic, and future disease risk.
To date, most epigenome-wide association studies (EWAS) of arsenic exposure have small sample sizes and used different data processing and analytical methods, making it difficult to compare results. The team sought to overcome this limitation by combining EWAS. These studies use genome-wide assays of epigenetic marks, such as DNAm, to identify associations between exposures and epigenetic variation.
Increasing Statistical Power
The team, led by trainee Anne Bozack, Ph.D., used existing SRP-funded datasets with information on DNAm. Researchers from Columbia provided data from participants in the Health Effects of Arsenic Longitudinal Study in Bangladesh who had high and low exposure to arsenic from drinking water as adults. UC Berkeley’s data were collected from adults in Northern Chile who were exposed to arsenic prenatally and in early life.
Epigenetic data was generated using microarray platforms on DNA collected from blood cells important in the immune response and buccal cells collected from a cheek swab. A microarray is a laboratory tool used to detect the expression of thousands of genes at the same time.
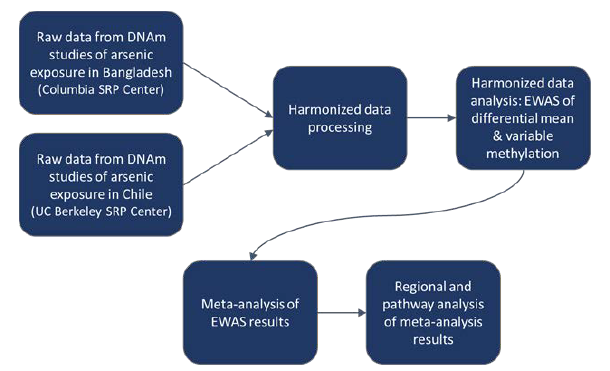
The researchers created a harmonized data processing and analysis pipeline, including establishing a consistent classification of exposure across datasets and data preprocessing steps to facilitate integration. Preprocessing included data normalization, quality control, and cleaning. They worked collaboratively via GitHub to standardize these steps across SRP centers.
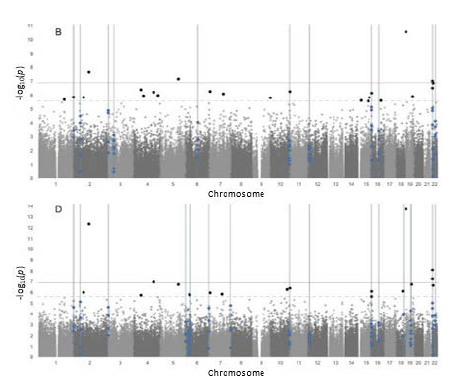
First, the Columbia and UC Berkeley researchers conducted analyses separately on their individual EWAS datasets but did not find any common arsenic-related DNAm signatures. However, when EWAS data was combined using their harmonized data processing and analysis pipeline, their meta-analysis revealed associations between arsenic exposure and variability in DNA methylation. Specifically, they identified 11 differentially variable regions within chromosomes among blood cells and 19 differentially variable regions when looking at both blood cells and buccal cells. Nine of these regions were common to both analyses. The researchers explained that genomic regions with increased variability in methylation have been associated with functional control of gene expression and may be particularly responsive to environmental conditions.
Using a Kyoto Encyclopedia of Genes and Genomes, or KEGG, pathway analysis to understand the biological functions of these signatures, the team identified several changes to important biological pathways known to be related to arsenic exposure. For example, one of the pathways identified was one carbon metabolism. One carbon metabolism plays a role in arsenic metabolism by facilitating urinary excretion and reducing arsenic toxicity. They also found changes related to autophagy, an important process of degrading unnecessary or damaged components within the cell to maintain normal function. Autophagy is a potential mechanism through which arsenic exposure may lead to health problems such as type 2 diabetes.
Future Directions
The researchers explained this study is the first to investigate associations between chronic arsenic exposure and differences in DNAm variability, which may serve as an important biological mechanism or biomarker of arsenic exposure.
According to the authors, this work provides a model for standardizing data analysis to leverage EWAS and identify other biologically meaningful pathways as markers of environmental exposures. As the first meta-analysis of DNAm and arsenic exposure, this study may also facilitate larger meta-analyses using EWAS with larger samples sizes. This would enable including more diverse populations and exposure levels or serve as a foundation for a validation study conducted in a larger cohort.
Their complete data processing and analysis pipeline is publicly available on GitHub and on an Open Science Framework (OSF) repository as a resource to other groups interested in combining data sets and data sharing. This resource could facilitate comparison with other EWAS and advance understanding of arsenic toxicity.
For More Information Contact:
Andres Cardenas
Stanford University
Department of Epidemiology and Population Health
Research Park
Stanford, California 94305-5405
Phone: 650-497-2815
Email: andresca@stanford.edu
Mary V Gamble
Columbia University
Environmental Health Sciences
Mailman School of Public Health
New York, New York 10032
Phone: 212-305-7949
Email: mvg7@columbia.edu
To learn more about this research, please refer to the following sources:
- Bozack AK, Boileau P, Wei L, Hubbard AE, Sille FC, Ferreccio Readi C, Acevedo J, Hou L, Ilievski V, Steinmaus CM, Smith MT, Navas-Acien A, Gamble MV, Cardenas A. 2021. Exposure to arsenic at different life-stages and DNA methylation meta-analysis in buccal cells and leukocytes. Environ Health 20:79. doi:10.1186/s12940-021-00754-7 PMID:34243768 PMCID:PMC8272372
To receive monthly mailings of the Research Briefs, send your email address to srpinfo@niehs.nih.gov.


