Superfund Research Program
Combining Analytical Chemistry and Machine Learning to Detangle Mixtures
View Research Brief as PDF(431KB)
Release Date: 02/01/2023
![]() subscribe/listen via iTunes, download(7.4MB), Transcript(90KB)
subscribe/listen via iTunes, download(7.4MB), Transcript(90KB)
NIEHS Superfund Research Program (SRP)-funded researchers demonstrated a significant step toward identifying individual chemical components in complex mixtures. Their approach uses advanced analytical techniques and sophisticated machine learning approaches while overcoming the time-consuming separation steps that preceded traditional chemical analysis.
Naomi Halas, Ph.D., Rice University; Ankit Patel, Ph.D., Baylor College of Medicine and Rice University; and Peter Nordlander, Ph.D., Rice University, led the team. The researchers developed a new strategy to identify individual polycyclic aromatic hydrocarbon (PAH) compounds in complex multicomponent mixtures. PAHs are a family of pollutants composed of fused benzene rings generated during incomplete combustion. PAHs and their metabolites — substances produced as the contaminants break down in the body or the environment — are known carcinogens, but often occur in the environment as complex mixtures, which makes detecting and identifying them difficult.
Combining Tools
The team’s strategy combines the ultrasensitive molecular fingerprinting capabilities of surface-enhanced Raman spectroscopy (SERS) with the complex signal separation and detection capabilities of machine learning. SERS is a type of chemical analysis based on chemical bonds' interaction with light. The resulting spectra of wavelengths is used to infer chemical structures.
Their machine learning method, Characteristic Peak Extraction (CaPE), can be used with SERS in an approach they called computational chromatography. In short, they used algorithms that de-mix SERS spectra from complex mixtures into individual PAH spectra and applied CaPE to extract characteristic SERS spectra based on the count and locations of detected peaks. Importantly, this approach helps address components of mixtures that occur at low concentrations and filters out noisy, or meaningless, data by ignoring non-characteristic peaks and focusing on those that are most unique within the dataset, creating a compressed, or simplified molecular fingerprint. Their strategy can also handle small frequency shifts, common to SERS, which confound other approaches.
To evaluate their method, the team collected SERS spectra for six different two-component PAH mixtures involving anthracene, pyrene, benzo(a)pyrene (BaP), and benz(a)anthracene. They also used more complex mixtures of all four PAHs at different concentration ratios.
They compared several common de-mixing algorithms with and without CaPE to see which performed best at pulling out individual chemicals, called de-mixed components. Then, the scientists used another algorithm to match unknown de-mixed components to specific PAHs in a PAH spectra library based on characteristic spectral features.
Getting Better Results
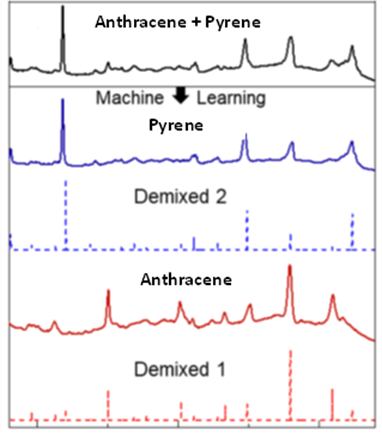
The research team aimed to observe how their approach performed with PAH mixtures of different complexity. In mixtures of two PAHs, their approach performed best for the anthracene and pyrene mixture and the anthracene and benzo(a)anthracene mixture, although in some cases minor features in the SERS spectra were not present in the resulting de-mixed components. Analysis of other mixtures contained some errors, however, the team explained that none prevented the de-mixed components from being matched visually or computationally to the correct PAH.
In more complex mixtures, with four PAHs, the de-mixed components did not match as closely with the SERS spectra. The best results were obtained for pyrene where the five most intense peaks matched the most intense peaks from SERS well. Despite the slightly less robust performance, their matching algorithm was still able to pair the de-mixed components to the correct PAHs.
When comparing different de-mixing methods with and without CaPE, they found that adding CaPE consistently improved performance, particularly for the most difficult mixtures like BaP with anthracene and the four-component mixtures. In particular, existing methods only matched about half of the correct PAHs, whereas CaPE extracted and assigned characteristic peaks effectively.
![Bar charts reflecting CaPE's rising capabilites when compared to other de-mixing methods. Steadily, the algorithm grew more successful against five de-mixing methods, in ascending order of: 4 PAH, B[a]P+ANTH, B[a]A+B[a]P, B[a]A+PYR, B[a]A+ANTH, PYR+B[a]P, PYR+ANTH.](https://connect.niehs.nih.gov/srp/1/ResearchBriefs/RB338_img3_no_caption.png)
According to the team, their results show that combining SERS and machine learning can more accurately, rapidly, and effectively de-mix compounds so that the individual chemicals can then be identified. This strategy can be used for rapidly detecting and identifying diverse chemicals in complex mixtures based on key molecular structures with no prior knowledge of their identity, the authors noted.
The data used in this study is available for download:
- Bajomo, Mary M., Ju, Yilong, Zhou, Jingyi, Elefterescu, Simina, Farr, Corbin, Zhao, Yiping, Neumann, Oara, Nordlander, Peter, Patel, Ankit, & Halas, Naomi J. (2022). Computational Chromatography: A Machine Learning Strategy for Demixing Individual Chemical Components in Complex Mixtures (Version 1) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.7406328
For More Information Contact:
Naomi Halas
Rice University
6100 Main Street
MS 378
Houston, Texas 77005
Phone: 713-348-5612
Email: halas@rice.edu
To learn more about this research, please refer to the following sources:
- Bajomo MM, Ju Y, Zhou J, Elefterescu S, Farr C, Zhao Y, Neumann O, Nordlander P, Patel AB, Halas N. 2022. Computational chromatography: a machine learning strategy for demixing individual chemical components in complex mixtures. Proc Natl Acad Sci U S A 119(52):e2211406119. doi:10.1073/pnas.2211406119 PMID:36534806
To receive monthly mailings of the Research Briefs, send your email address to srpinfo@niehs.nih.gov.


