Superfund Research Program
Mapping Microbe Interactions That Support PCB-Degrading Bacteria
Release Date: 01/10/2024
![]() subscribe/listen via iTunes, download(6.0MB), Transcript(107KB)
subscribe/listen via iTunes, download(6.0MB), Transcript(107KB)
Researchers partially funded by the NIEHS Superfund Research Program (SRP) mapped interactions between microbes that may support the growth of certain bacteria that degrade polychlorinated biphenyls (PCBs), a harmful contaminant. By harnessing those microbial relationships, researchers could improve the bioremediation, or bacterial breakdown, of PCBs from the environment, according to a team led by Timothy Mattes, Ph.D., University of Iowa SRP Center.
One kind of bacteria that is used to clean up PCBs is Dehalococcoides, which is highly specialized and relies on interactions with other bacteria to thrive. Dehalococcoides produces a special enzyme known as reductive dehalogenase, and this enzyme breaks down PCBs.
For their study, the researchers used sophisticated genetic techniques to identify microbial genes that could support growth of Dehalococcoides. Their approach is part of a field called metagenomics, which explores the structure and function of DNA that appears in a bulk sample of organisms.
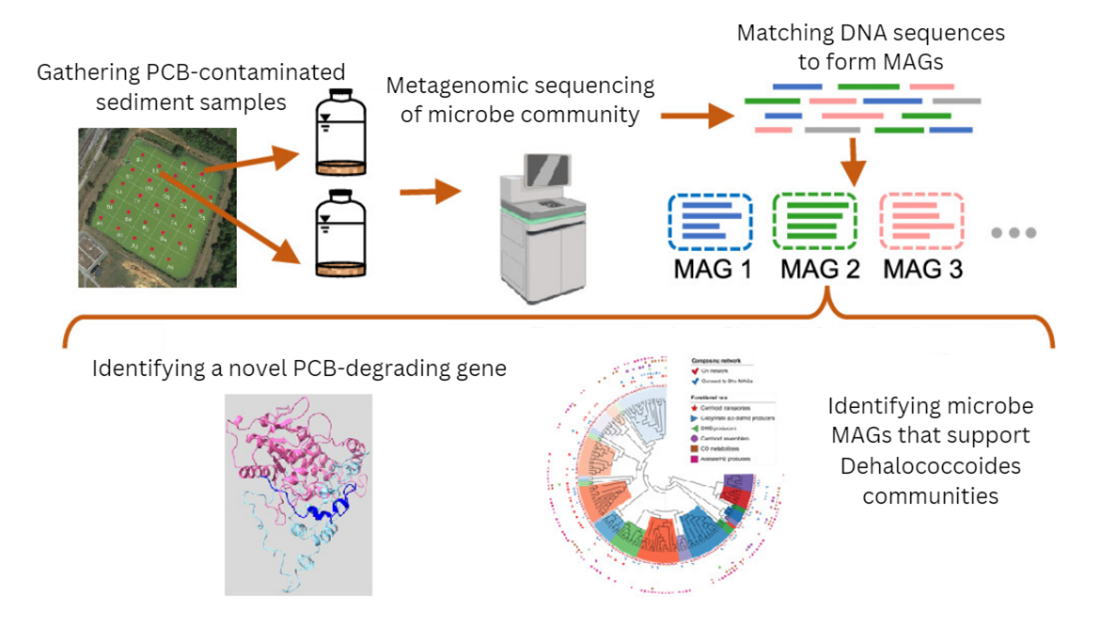
Growing and Sequencing Microbial Communities
The team first obtained PCB-contaminated sediment samples that contained PCB-degrading bacteria and created four microcosms, or miniature environments, with the sediment samples. The researchers incubated the microcosms for 200 days, and sequenced DNA and RNA segments for analysis at the end of the incubation.
Next, the team pieced together the short DNA sequences using a computer program, essentially reconstructing the microbial genomes found in each microcosm. In total, they produced 160 of these metagenome assembled genomes (MAGs), including three from Dehalococcoides bacteria.
The scientists classified the MAGs into 24 distinct high-level groups, including Dehalococcoides. The researchers then analyzed how gene expression from different MAGs might correlate with each other, and whether those interactions influence Dehalococcoides. The idea was to see if certain MAGs supported Dehalococcoides growth and PCB degradation activity, and which bacteria the supporting MAGs belonged to.
Linking PCB-Degrading and Supporting Bacteria
The researchers found that 112 MAGs contained genes to generate metabolic byproducts, like hydrogen, which were related to Dehalococcoides growth. Of those, the team identified 31 MAGs that expressed genes at the same time as the Dehalococcoides MAGs. The findings suggest that the activity of microbes that produce more of those factors could improve bioremediation by Dehalococcoides, according to the researchers.
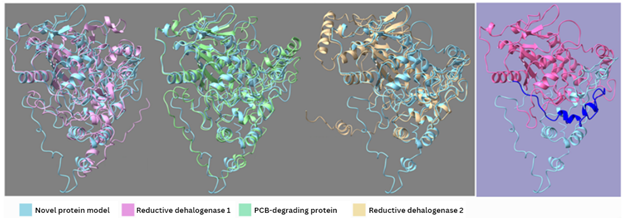
When examining the Dehalococcoides MAGs, the researchers also discovered a novel reductive dehalogenase gene related to PCB biodegradation and compared its predicted protein structure to other similar genes in the National Center for Biotechnology Information database. Only limited sections of the predicted protein structure of the novel gene were similar to other reductive dehalogenases, including one PCB-degrading gene.
The researchers believe these insights into microbial genome interactions may guide efforts to enhance the efficiency of PCB-degrading bacteria. The study could also facilitate new methods for improving bioremediation of PCB by changing the amount and type of microbes in a desired area.
For More Information Contact:
Timothy E Mattes
University of Iowa
4112 Seamans Center for the Engineering Arts and Sciences
Iowa City, Iowa 52242
Phone: 319-335-5065
Email: tim-mattes@uiowa.edu
To learn more about this research, please refer to the following sources:
- Dang H, Ewald JM, Mattes TE. 2023. Genome-resolved metagenomics and metatranscriptomics reveal insights into the ecology and metabolism of anaerobic microbial communities in PCB-contaminated sediments. Environ Sci Technol 57(43):16386-16398. doi:10.1021/acs.est.3c05439 PMID:37856784 PMCID:PMC10621002
To receive monthly mailings of the Research Briefs, send your email address to srpinfo@niehs.nih.gov.


