Superfund Research Program
New Method Quickly Screens Chemicals for Cancer Risk
View Research Brief as PDF(418KB)
Release Date: 06/05/2019
![]() subscribe/listen via iTunes, download(7.3MB), Transcript(87KB)
subscribe/listen via iTunes, download(7.3MB), Transcript(87KB)
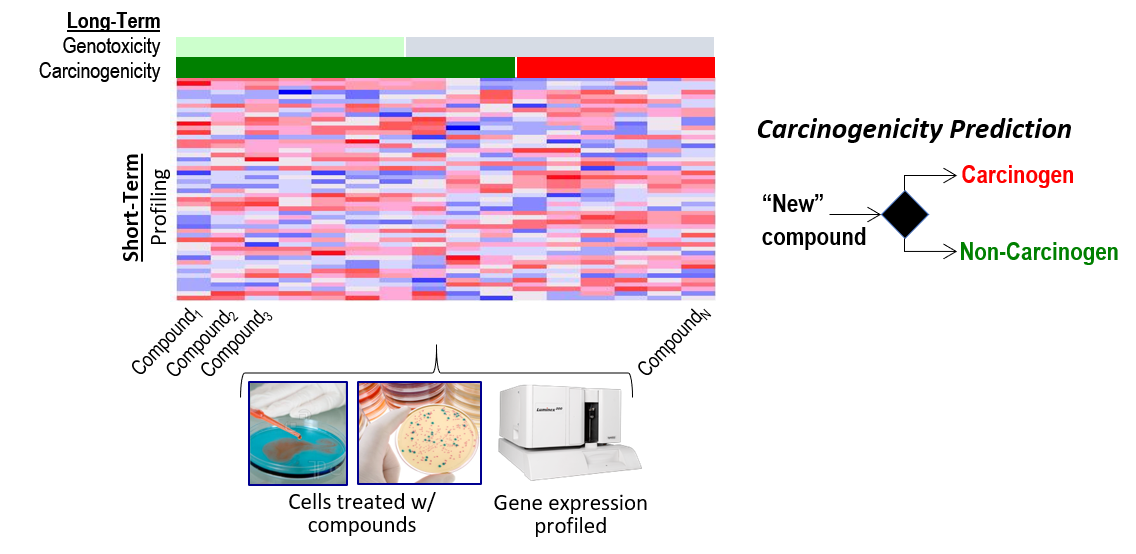
Boston University (BU) researchers, in collaboration with researchers at the National Toxicology Program (NTP) and the Broad Institute, have developed and evaluated a new approach to assess whether exposure to a chemical increases a person’s long-term cancer risk. The fast, cost-effective method uses gene expression profiling, which measures the activity of a thousand or more genes to capture what is happening in a cell. Based on gene expression profiling data, the researchers were able to infer specific biological changes at the cellular level and predict potential carcinogenicity of chemicals, or the ability of chemicals to cause cancer.
Of the tens of thousands of chemicals in commercial use, less than two percent have been thoroughly tested for their potential carcinogenicity, in part because the current chemical screening process is costly and time-consuming. Led by Stefano Monti, Ph.D., of the BU Superfund Research Program, the team set out to develop a short-term cell-based screening process to predict long-term cancer risk. According to the authors, their method provides a promising solution that could be used to prioritize chemicals for further cancer testing.
Developing Computer Models to Find Gene Expression Patterns
The researchers exposed human liver cell lines to hundreds of individual chemicals known to be carcinogens and noncarcinogens and measured changes in gene expression within the cells using the Luminex L1000 platform. L1000 measures the expression of nearly 1,000 landmark genes, those determined to best predict the expression levels of the remaining genes in the transcriptome, or all RNA molecules in that cell.
Then, they fed the resulting data to a computer model that used machine learning techniques to find patterns within the gene expression profiles that would distinguish carcinogenic from non-carcinogenic chemicals.
These patterns then formed the basis of the model that researchers used to predict the long-term carcinogenicity of a variety of different chemicals. The model was evaluated by testing its accuracy on chemicals already known to induce or not induce cancer, using data available from DrugMatrix, Connectivity Map (CMAP), and Toxicology in the 21st Century (Tox21).
Across the entire dataset, the model did not perform well. However, when they narrowed it down to chemicals with high biological activity levels, focusing on the 63 chemicals with the highest levels of bioactivity, they found that their model accurately predicted carcinogenicity, as well as genotoxicity, or damage to the cell’s genetic material.
They found that many of the expression profiles in their study involved genes implicated in DNA damage and repair processes, as well as immune-related pathways, metabolism pathways, and cell communication. Comparing these gene expression profile differences between carcinogens and noncarcinogens may inform further research on how different compounds can lead cells to become cancerous.
The research team made their data, including 6,000 gene expression profiles of more than 300 liver carcinogens and noncarcinogens, available to other researchers. They also created a portal for the public to search and visualize the results.
Alternative Methods for Safety Testing
The gold standard of carcinogenicity testing, the two-year rodent bioassay, is time-consuming and can require up to $4 million and more than 800 animals for every compound evaluated. The new approach represents a step forward for Tox21, a federal program focused on developing new ways to rapidly test whether chemicals may affect humans and the environment.
Although more work needs to be done to optimize and validate this approach before it can be applied in regulatory and clinical settings, it has the potential to provide a fast and cost-effective means to screen chemicals for further testing, according to the authors. Additionally, this approach could be extended to evaluate other negative effects of exposure, such as endocrine and metabolic disruption.
For More Information Contact:
Stefano Monti
Boston University
75 E. Newton St.
Boston, Massachusetts 02118
Phone: 617-414-7031
Email: smonti@bu.edu
To learn more about this research, please refer to the following sources:
- Li A, Lu X, Natoli T, Bittker J, Sipes N, Subramanian A, Auerbach SS, Sherr DH, Monti S. 2019. The Carcinogenome Project: in vitro gene expression profiling of chemical perturbations to predict long-term carcinogenicity. Environ Health Perspect 127:47002. doi:10.1289/EHP3986 PMID:30964323 PMCID:PMC6785232
To receive monthly mailings of the Research Briefs, send your email address to srpinfo@niehs.nih.gov.


