Superfund Research Program
June 2023
By Julie Leibach
Researchers with the NIEHS-funded Massachusetts Institute of Technology (MIT) Superfund Research Program (SRP) Center have found distinctive genetic patterns signifying damage from the toxic chemical N-nitrosodimethylamine, or NDMA. The patterns could potentially be used to monitor cancer development and inform therapeutic interventions.
The International Agency for Research on Cancer classifies NDMA as likely to cause cancer in humans. For example, in 2021 state health officials reported an association between prenatal NDMA exposure and childhood cancer among kids living in Wilmington, Massachusetts, in the 1990s. The conduit was drinking water tainted by activities from a former chemical manufacturing facility, now the location of a Superfund site.
Once used to make rocket fuel, NDMA is no longer commercially available. However, industrial processes still produce NDMA as a byproduct, and the chemical remains a public health concern near Superfund sites.
“Superfund sites tend to disproportionately affect people of color and low-income communities,” said co-first author and SRP trainee Amanda Armijo, D.V.M., Ph.D. “Our study not only investigates the origins of certain cancers but also provides background for important issues of environmental justice.”
Seeing a Sawtooth
For their study, the MIT researchers wondered if they could identify signs of NDMA exposure in DNA.
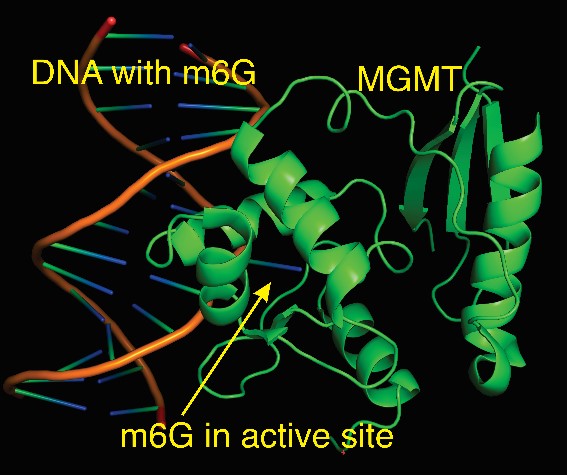
Upon exposure, the body breaks down NDMA into fragments that can harm DNA. One of those fragments reacts with DNA to form a complex called m6G. That complex can cause an error in DNA base-pairing, a process by which one DNA strand connects to another through molecules called bases to form a two-stranded helix.
During typical base-pairing, the molecule adenine (A) pairs with thymine (T), and cytosine (C) complements guanine (G). However, m6G causes G to mistakenly pair with T. In normally functioning cells, a DNA repair protein would prevent the error by undoing the m6G complex. Otherwise, the newly introduced T will pair with A during DNA replication. Future replication will perpetuate that mutation, or change, in the genetic sequence.
To explore NDMA-associated mutations, the researchers first treated mice with the chemical. Ten weeks later, they applied a highly sensitive DNA sequencing tool to cells from mouse lungs and livers, organs targeted by NDMA.
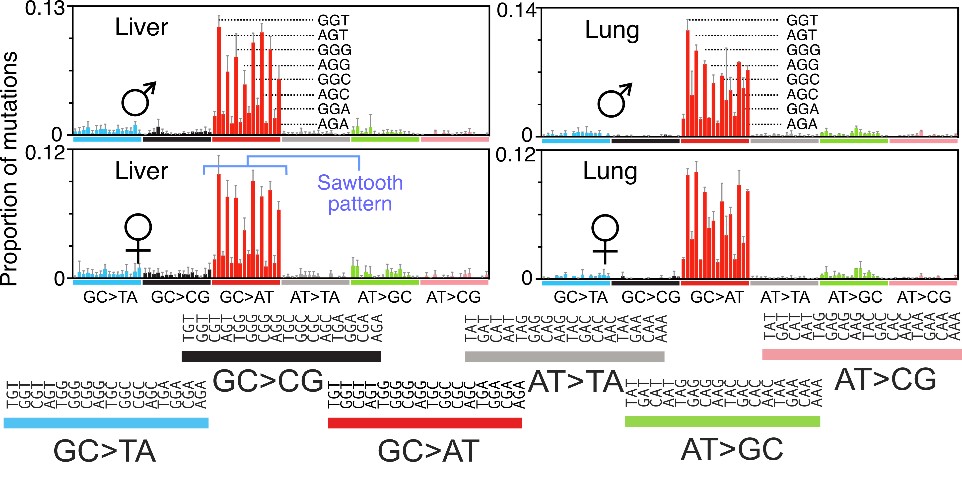
They identified a unique pattern of mutations showing that, where G-C pairs should have been, A-T pairs appeared. Plotted in a bar graph, the pattern resembled the jagged edges of a saw. In addition, the mutations occurred only in small sections of DNA with specific base sequences, indicating that m6G acts discriminately.
In a separate experiment, the team analyzed DNA in exposed mice lacking MGMT, the DNA repair protein that undoes the m6G complex. They found even more mutations in virtually the same pattern, confirming the protective role of intact MGMT.
Unpacking a Paradox
The team was also interested in whether anticancer drugs known as alkylating agents — a chemical class that includes NDMA — cause similar mutational patterns. Like NDMA, these drugs interfere with proper base-pairing. Their success at thwarting cancer hinges on a different set of DNA repair proteins that destroy mutated DNA, eventually leading to cell death.
In one experiment, the researchers exposed cells to several anticancer drugs and sequenced the resulting mutations. In addition, they consulted an existing database of mutations documented in cancer patients treated with one of the drugs. Results indicated that the cancer therapies indeed caused mutational patterns like those induced by NDMA.
The findings offer insight into a longstanding puzzle as to why some cancer therapies can both treat and cause tumors, according to the researchers.
“This is somewhat speculative, but cancer cells that survive initial treatment by these drugs may lack the DNA processing mechanisms that cause cells to die. Or, they may lack the ability to prevent the base-pairing error caused by m6G,” explained lead author John Essigmann, Ph.D., with the MIT SRP Center. “For either or both reasons, the base-pairing error persists as cells divide, potentially creating a relapsing and harder-to-treat tumor. If we were to sequence that tumor, we believe we would see the sawtooth pattern.”
The distinct mutational pattern associated with exposure to NDMA and other alkylating agents could be used as a biomarker, or sign, of early cancer development, the researchers noted. Identifying those signals early on could prompt surgery to remove small tumors before they evolve into larger, more problematic malignancies, according to the team.


